Zweigerdt Lab
Applying human pluripotent stem cells (hPSCs), we are working on aspects of developmental biology and regenerative medicine focused on the heart as well other tissues and lineages such as macrophages. The research is structured into separate but closely interconnected topics and projects:
1. Bioprocess development for the clinical translation of hPSC progenies, particularly of mesendoderm derivatives such as cardiomyocytes, endothelial cells, macrophages and definitive endoderm.
2. Mechanisms of mesendoderm specification and heart development, such as the in vitro induction, characterization, and stabilization of cardiac progenitors (SCPs) and recapitulating early steps of native heart development in vitro via Heart-Forming Organoids (HFOs), which comprises the functional interplay of heart anlagen with foregut endoderm.
3. Modeling (congenital) heart diseases in a dish. Our projects in this field include electrophysiological disorders (e.g. Brugada Syndrome due to mutations in the SCN5A gene), disruption of cardiac transcription factors (e.g. NKX2.5 Knock Out in HFOs), and studying cardiac hypertrophy in hPSC-cardiomyocytes based on pharmacological stimuli or genetic mutations (e.g. in MYH7 or MYBPC3).
Collaboration with Industry: Besides academic partners (within and beyond the MHH; listed in relation to specific projects below) many of our research projects involve the collaboration with Industry Partners including DASGIP-Eppendorf, LaVision BioTec, Miltenyi Biotec, Novo Nordisk, Thermo Fisher, and Thinking Research Instruments (listed in alphabetic order).
1. Bioprocess development for the clinical translation of hPSC progenies
In recent years, several hPSC-based therapy concepts have progressed towards first in-man studies. This progress is accompanied with the need for efficient and robust culture and differentiation strategies to produce hPSCs at clinically relevant quantities, quality and at commercially viable conditions.
Our vision for the clinical and industry compliant production of hPSC progenies is centered on the use of stirred tank bioreactor (STBR) technologies to ensure robustness, scalability, automation, and ultimately GMP-compliant cell production processes. This concept includes the culture, mass production and differentiation of hPSCs as entirely matrix-free, cell-only aggregates in stirred suspension culture. The strategy enables rational process optimization by in silico modeling, upscaling by a seed-train approach, and closed-loop manufacturing required for GMP compliance and automation. (Figure 1).
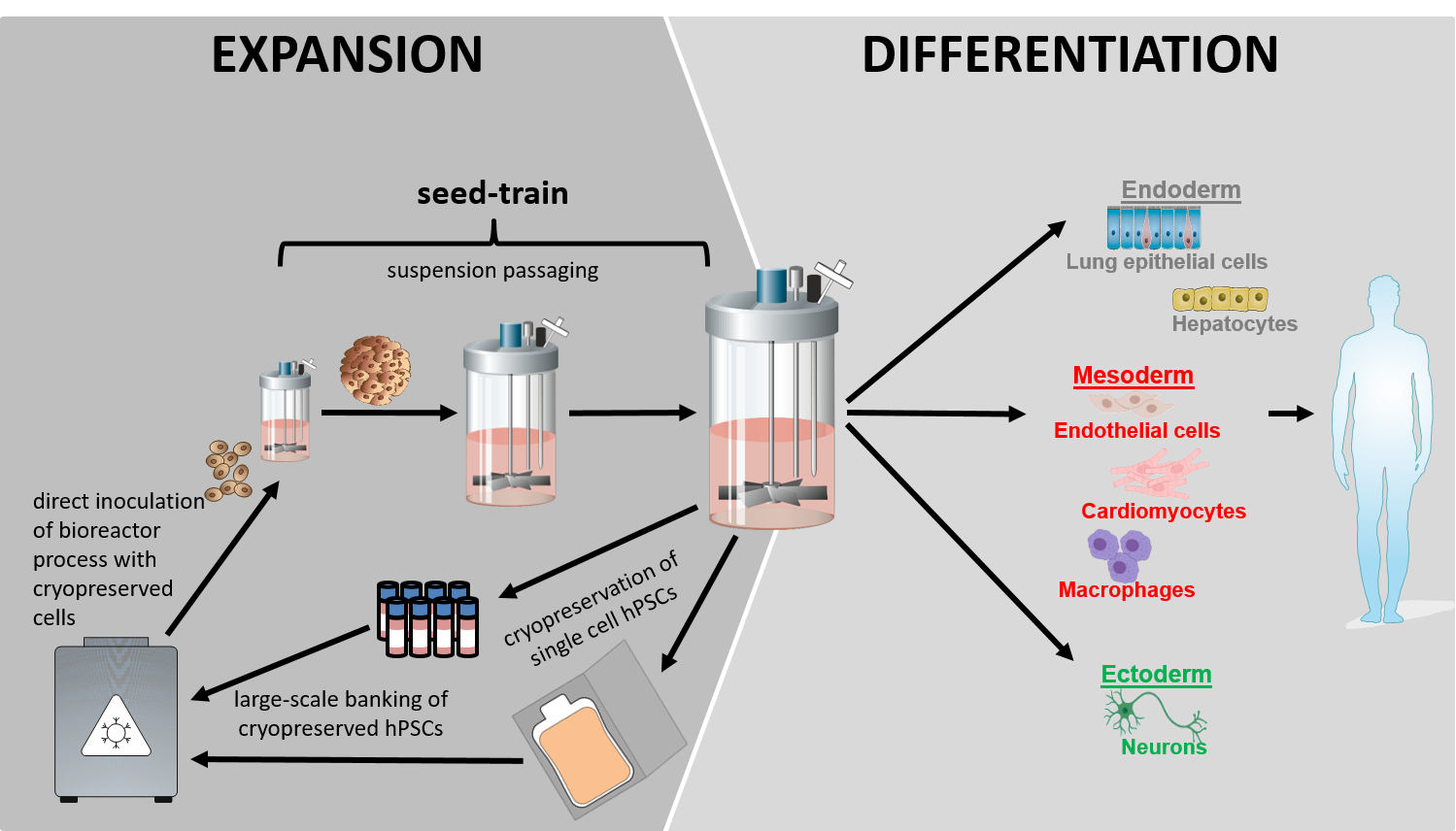
Current developments:
- Generation of cryopreserved cell banks for direct suspension culture inoculation in STBRS thereby enabling entirely matrix-free hPSC culture.
- Promoting process efficiency by in silico process modeling both for hPSC expansion and lineage-specific differentiation.
- Advancing the rational of cardiomyogenic differentiation and in-process control by secretome analysis.
- Enabling closed-loop bioprocessing for GMP-compliant cell production.
Collaborations and projects on Bioprocess development:
On the one hand, we are focusing on the upscaling and optimization of processes for pluripotent cell aggregates production, serving as “the raw material” for linage-directed differentiation(s).
Our second focus is on the directed cardiomyogenic differentiation of hPSCs. In cooperation with the MHH groups of Ulrich Martin (LEBAO), Axel Haverich (HTTG) and numerous other investigators, the process-derived cardiomyocytes are evaluated for clinical heart repair in the iCARE project (LINK). The application of hPSC-derived cardiomyocytes and endothelial cells - the latter being generated by the group of Ruth Olmer (MHH/ LEBAO) - is performed in collaboration with the LEBAO groups of Ina Gruh, Andres Hilfiker/ Birgit Andree, Andreas Martens, and Thomas Aper in the 3D-Heart-2B project, aiming at the in vitro engineering of multilayered, vascularized, linear heart-tube tissue construct (LINK).
Another important collaboration aims at the continuous large scale production of hiPSC-macrophages in close collaboration with the group of Nico Lachmann (MHH) which includes funding by the industrial partner Novo Nordisk (Denmark; https://www.mhh.de/en/presse/mhh-insight/news-detailed-view/cells-of-the-future).
- Diana Massai (Politecnico di Torino, Italy)
- Elizabeth Lipke and Selen Cremaschi (Auburn University, US)
- DASGIP-Eppendorf (Hamburg, Germany)
- Thermo Fisher Scientific (Darmstadt, Germany)
- Thinking Research Instruments (Hamburg, Germany)
- Ackermann et al. Nat Protoc 2022 (https://www.nature.com/articles/s41596-021-00654-7 ),
- Manstein et al. STAR Protoc. 2021 (https://star-protocols.cell.com/protocols/1227 ),
- Manstein et al. Stem Cells Transl Med. 2021 (https://stemcellsjournals.onlinelibrary.wiley.com/doi/full/10.1002/sctm.20-0453 ),
- Williams et al. Front Bioeng Biotechnol 2020 (https://www.frontiersin.org/articles/10.3389/fbioe.2020.00851/full ),
- Halloin et al. Stem Cell Reports 2019 (https://www.sciencedirect.com/science/article/pii/S2213671119303303?via%3Dihub ),
- Sahabian et al. Cells 2019 (https://www.mdpi.com/2073-4409/8/12/1571 ),
- Ackermann et al. Nat Commun. 2018 (https://www.nature.com/articles/s41467-018-07570-7 ),
- Olmer et al. Stem Cell Reports 2018 (https://www.sciencedirect.com/science/article/pii/S2213671118301449?via%3Dihub ),
- Massai et al Sci Rep. 2017 (https://www.nature.com/articles/s41598-017-04158-x ),
- Kropp et al. Stem Cells Transl Med. 2016 (https://stemcellsjournals.onlinelibrary.wiley.com/doi/full/10.5966/sctm.2015-0253 ),
- Kempf et al. Nat Protoc. 2015 (https://www.nature.com/articles/nprot.2015.089 ),
- Kempf et al. Stem Cell Reports. 2014 (https://www.sciencedirect.com/science/article/pii/S2213671114003026?via%3Dihub ),
- Zweigerdt et al. Nat Protoc. 2011 (https://www.nature.com/articles/nprot.2011.318 ).
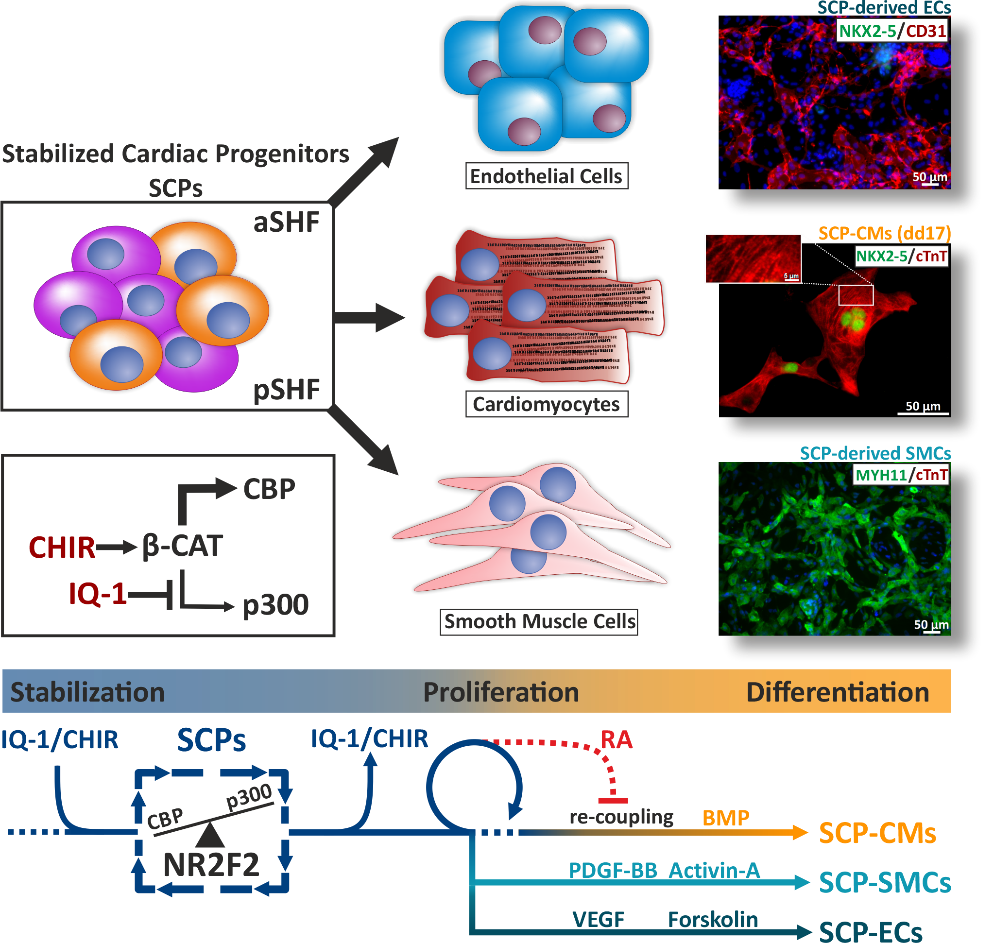
2. Mechanisms of mesendoderm specification, heart development and regeneration
Equivalent to embryogenesis, the in vitro transition from hPSC pluripotency towards lineage specification and differentiation is highly complex and poorly understood. However, recent work revealed that many of the complex molecular and cellular mechanisms controlling the native development of cells, tissues and organs in the embryo (often studied in model organisms such as frogs, fish and mice) can be closely recapitulated and studied by advanced strategies of hPSC differentiation in vitro, including so-called “organoids”. Focusing on mesendoderm specification and heart development, we are working on several projects in this exciting field of hPSC research.
Cardiac Progenitors only transiently exist during heart development, underlying the lack of regenerative capacity in the adult organ. Directed cardiac differentiation of hPSCs is a potent in vitro model of cardiogenesis. However, the transient nature of cardiac progenitor cells also hampers the in vitro investigation of mechanisms controlling their development. Histone Acetyl Transferases (HATs) have been shown to play a pivotal role in heart development and homeostasis. In spite of their ubiquitous role in gene networks regulation, systematic studies in heart development and diseases are missing. We are specifically interested in studying the effect of small molecular inhibitors capable of modulating HATs during heart development as a novel approach to manipulate hPSC-cardiac lineage specification and differentiation, particularly aiming at establishing Stabilized Cardiac Progenitors (SCPs), to investigate the molecular properties of this transiently occurring and intensively sought-after lineage.
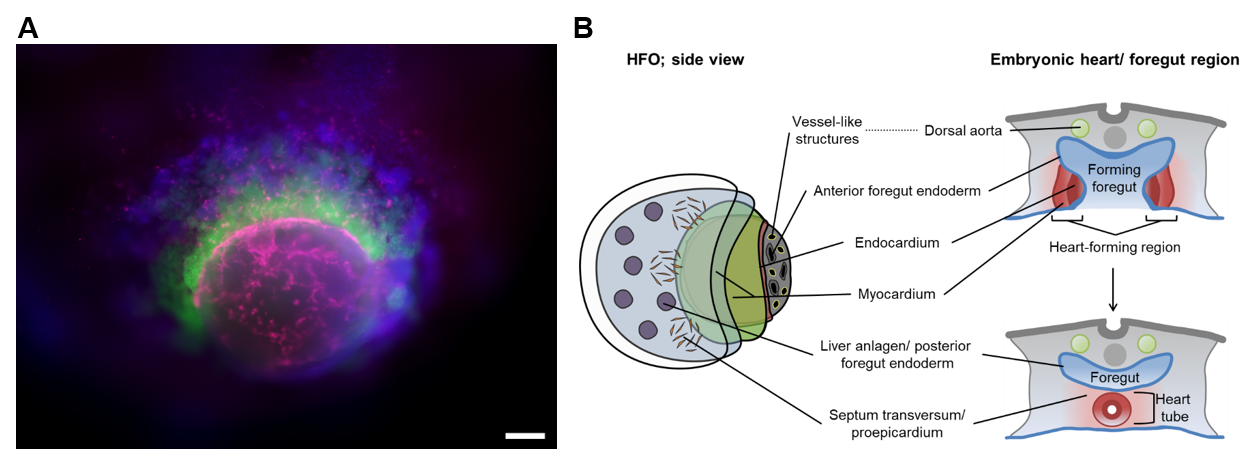
Heart-Forming Organoids (HFOs): In response to proper stimuli, hPSCs self-organize into 3D structures called "organoids" resembling embryo-like tissue patterns in vitro. We have for the first time developed a highly structured organoid model of early heart, foregut and vasculature development termed “heart-forming organoids” (HFOs). HFOs are composed of cardiomyocytes, endocardial-like cells, septum-transversum-like anlagen, anterior versus posterior foregut endoderm tissues and a vascular network (Figure 3). Besides deciphering complex mechanisms of human organogenesis in a dish, the HFO model can also be used for replacing animal studies in the process of drug discovery and development such as teratogenicity assessment or drug screenings in frame of congenital heart disease studies and tissue regeneration.
For the advanced assessment of complex 3D structures over time/ in real time (4D) and the distinct manipulation and tracking of individual cells, we have established projects with industrial leaders in microscopy i.e. LaVision BioTec/ Miltenyi Biotec (MoDiaNo) and academic specialists in femtosecond laser nanosurgery- (FLN-) based technologies (Kalies/ Heisterkamp, Leibniz University Hannover).
Stabilized Cardiac Progenitors:
- Dorothee Bornhorst (Boston Children’s Hospital, Harvard University, USA)
- Salim Abdelilah-Seyfried (MHH/ Potsdam University, Germany)
- Lavanya Muthukrishnan Iyer (Göttingen Medical University/ MDC Berlin; Germany)
- Laura C. Zelarayán-Behrend (Göttingen Medical University, Germany)
- Dorota Zawada, Alessandra Moretti (Munich Technical University, Germany)
- Alexander Gödel (Karolinska Institutet, Sweden)
- Michael Morgan, Lange Lucas, Axel Schambach (MHH)
- Andreas Leffler (MHH)
- Gergana Dobreva (Anatomy and Developmental Biology Medical Faculty Mannheim, Heidelberg)
Heart-forming organoids:
- Simone Liebscher, Katja Schenke-Layland (Eberhard Karls University Tübingen, Germany)
- Jan Hegermann (MHH, Germany)
- Lena Nolte, Heiko Meyer (Laser Center Hannover)
- Jeanne de la Roche, Stefan Thiemann, Christian Wahl-Schott (MHH, Germany)
- Stefan Kalies, Alexander Heisterkamp (Leibniz University Hannover)
- LaVision BioTec (Bielefeld, Germany)/ Miltenyi Biotec (Bergisch-Gladbach, Germany)
- Lothar Koch, Boris Chichkov (Leibniz University Hannover)
- Drakhlis L, Devadas SB, Zweigerdt R. Generation of heart-forming organoids from human pluripotent stem cells. Nat Protoc. 2021 Dec;16(12):5652-5672. doi: 10.1038/s41596-021-00629-8. Epub 2021 Nov 10. PMID: 34759383.
- Drakhlis L, Biswanath S, Farr CM, Lupanow V, Teske J, Ritzenhoff K, Franke A, Manstein F, Bolesani E, Kempf H, Liebscher S, Schenke-Layland K, Hegermann J, Nolte L, Meyer H, de la Roche J, Thiemann S, Wahl-Schott C, Martin U, Zweigerdt R. Human heart-forming organoids recapitulate early heart and foregut development. Nat Biotechnol. 2021 Feb 8. doi: 10.1038/s41587-021-00815-9. Epub ahead of print. PMID: 33558697.
- Müller D, Hagenah D, Biswanath S, Coffee M, Kampmann A, Zweigerdt R, Heisterkamp A, Kalies SMK. Femtosecond laser-based nanosurgery reveals the endogenous regeneration of single Z-discs including physiological consequences for cardiomyocytes. Sci Rep. 2019 Mar 6;9(1):3625. doi: 10.1038/s41598-019-40308-z.
- Gaspari E, Franke A, Robles-Diaz D, Zweigerdt R, Roeder I, Zerjatke T, Kempf H. Paracrine mechanisms in early differentiation of human pluripotent stem cells: Insights from a mathematical model. Stem Cell Res. 2018 Oct;32:1-7. doi: 10.1016/j.scr.2018.07.025. Epub 2018 Aug 18. PMID: 30145492.
- Koch L, Deiwick A, Franke A, Schwanke K, Haverich A, Zweigerdt R, Chichkov B.Laser bioprinting of human induced pluripotent stem cells-the effect of printing and biomaterials on cell survival, pluripotency, and differentiation. Biofabrication. 2018 Apr 25;10(3):035005. doi: 10.1088/1758-5090/aab981
- Kempf H, Olmer R, Haase A, Franke A, Bolesani E, Schwanke K, Robles-Diaz D, Coffee M, Göhring G, Dräger G, Pötz O, Joos T, Martinez-Hackert E, Haverich A, Buettner FFR, Martin U, Zweigerdt R. Bulk cell density and Wnt/TGFbeta signalling regulate mesendodermal patterning of human pluripotent stem cells. Nat Commun. 2016 Dec 9;7:13602. doi: 10.1038/ncomms13602. PMID: 27934856; PMCID: PMC5155150.
3. Modeling (congenital) heart diseases in a dish
hPSC-derived cardiomyocytes and cardiac progenitors hold great potential for both advanced cell therapies and in vitro modeling of cardiac diseases. Applying our technologies for the bioreactor-based mass production of hPSC-derived cardiomyocytes (which can be applied in any screening-compliant culture format) and the generation of more sophisticated heart-forming organoids (HFOs) described above, we are developing small- to mid-scale screening assays for modeling heart disease in a dish. Our successful examples of this approach include electrophysiological disorders (e.g. Brugada Syndrome due to mutations in the SCN5A gene), disruption of cardiac transcription factors (e.g. NKX2.5 knock-out in HFOs), and most recent investigations on cardiac hypertrophy in hPSC-derived cardiomyocytes based on pharmacological stimuli or genetic mutations e.g. in the MYH7 or MYBPC3 genetic loci (DFG project cMyBP-C-Mutationen FKZ: ZW 64/4-2). This research strongly depends on the expression patterns of sarcomeric proteins, which depends on numerous factor but in particular the culture format applied to hPSC-CMs after differentiation (Figure 3).
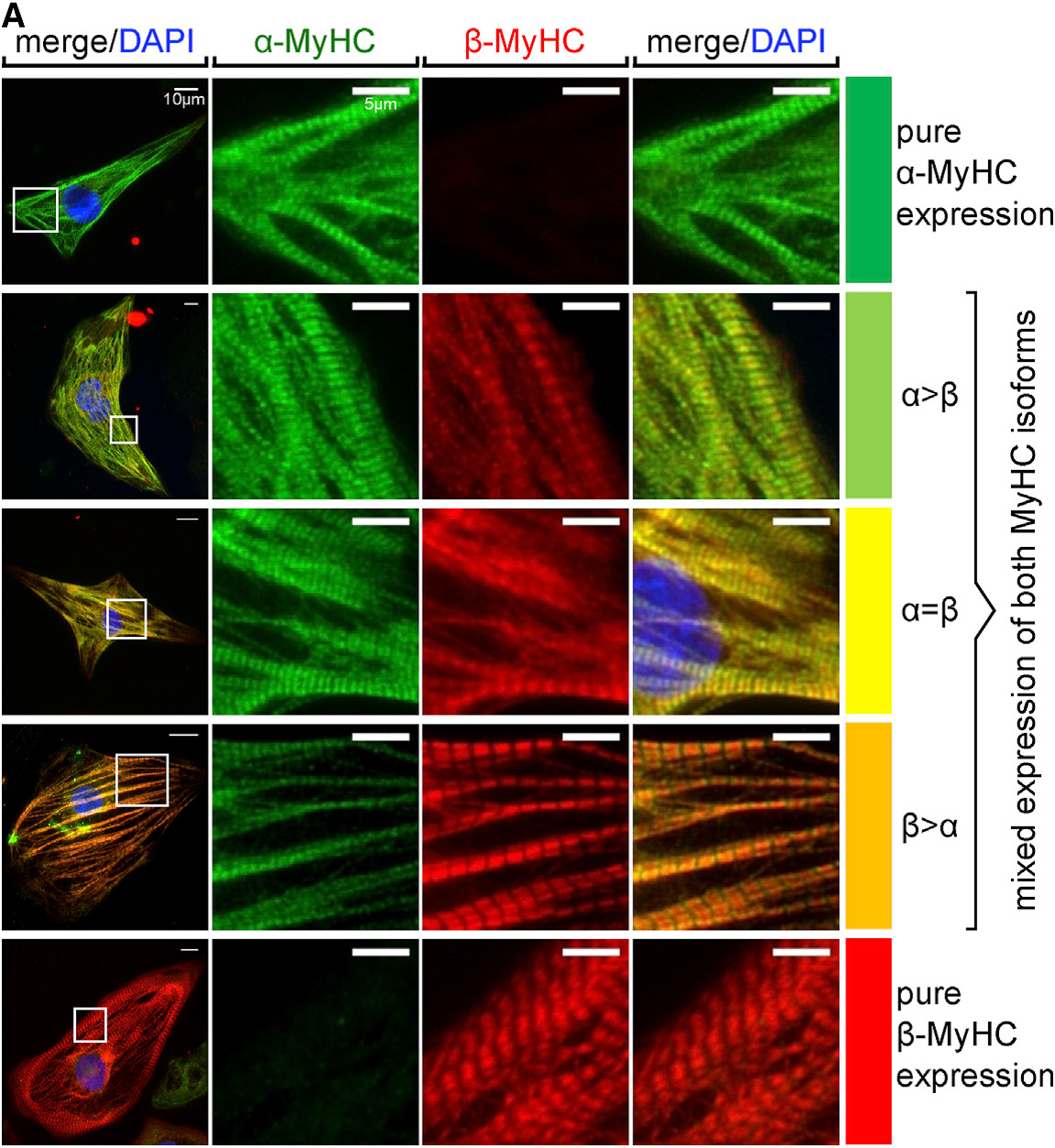
For developing a disease model with predictive potential, it is of the foremost importance to understand its limitations and biological boundaries. This is particularly true when trying to model complex pathologies such as cardiac hypertrophy. Prolonged mechanical overload on the heart or mutations of sarcomeric proteins may result in hypertrophic remodeling of the heart. However, to better understand the underlying mechanism in vitro, we are developing a 3D model of cardiac hypertrophy by using neurohormonal stimulation. This model could be further used for a mid-throughput screen to conduct trials on a number of drug candidates, which in turn could reduce the usage of animals in line with the 3R principles (Replacement, Reduction and Refinement).
- Christian Bär (MHH )
- Martin Fischer (MHH)
- Bogdan Iorga (MHH)
- Theresia Kraft (MHH )
- Manuel H. Taft (MHH)
- Dietmar J. Manstein (MHH)
- Thomas Thum (MHH)
- Natalie Weber (MHH)
- Kai C. Wollert (MHH )
- Chatterjee S, Hofer T, Costa A, Lu D, Batkai S, Gupta SK, Bolesani E, Zweigerdt R, Megias D, Streckfuss-Bömeke K, Brandenberger C, Thum T, Bär C. Mol Ther. Telomerase therapy attenuates cardiotoxic effects of doxorubicin. 2021 Apr 7;29(4):1395-1410. doi: 10.1016/j.ymthe.2020.12.035. Epub 2021 Jan 1
- Merkert S, Wunderlich S, Beier J, Franke A, Schwanke K, Göhring G, Kraft T, Francino A, Zweigerdt R, Martin U. Generation of two iPSC clones (MHHi021-A and MHHi021-B) from a patient with hypertrophic cardiomyopathy with p.Arg723Gly mutation in the MYH7 gene. Stem Cell Res. 2021 Apr;52:102208. doi: 10.1016/j.scr.2021.102208. Epub 2021 Feb 3. PMID: 33578365.
- Latham SL, Weiß N, Schwanke K, Thiel C, Croucher DR, Zweigerdt R, Manstein DJ, Taft MH. Myosin-18B Regulates Higher-Order Organization of the Cardiac Sarcomere through Thin Filament Cross-Linking and Thick Filament Dynamics. Cell Rep. 2020 Sep 1;32(9):108090. doi: 10.1016/j.celrep.2020.108090.
- Hunkler HJ, Hoepfner J, Huang CK, Chatterjee S, Jara-Avaca M, Gruh I, Bolesani E, Zweigerdt R, Thum T, Bär C. The Long Non-coding RNA Cyrano Is Dispensable for Pluripotency of Murine and Human Pluripotent Stem Cells. Stem Cell Reports. 2020 Jul 14;15(1):13-21. doi: 10.1016/j.stemcr.2020.05.011. Epub 2020 Jun 11. PMID: 32531193
- Weber N, Kowalski K, Holler T, Radocaj A, Fischer M, Thiemann S, de la Roche J, Schwanke K, Piep B, Peschel N, Krumm U, Lingk A, Wendland M, Greten S, Schmitto JD, Ismail I, Warnecke G, Zywietz U, Chichkov B, Meißner J, Haverich A, Martin U, Brenner B, Zweigerdt R, Kraft T. Advanced Single-Cell Mapping Reveals that in hESC Cardiomyocytes Contraction Kinetics and Action Potential Are Independent of Myosin Isoform. Stem Cell Reports. 2020 May 12;14(5):788-802. doi: 10.1016/j.stemcr.2020.03.015. Epub 2020 Apr 16. PMID: 32302556; PMCID: PMC7220955.
- de la Roche J, Angsutararux P, Kempf H, Janan M, Bolesani E, Thiemann S, Wojciechowski D, Coffee M, Franke A, Schwanke K, Leffler A, Luanpitpong S, Issaragrisil S, Fischer M, Zweigerdt R. Comparing human iPSC-cardiomyocytes versus HEK293T cells unveils disease-causing effects of Brugada mutation A735V of NaV1.5 sodium channels. Sci Rep. 2019 Aug 1;9(1):11173. doi: 10.1038/s41598-019-47632-4. PMID: 31371804; PMCID: PMC6673693.
- Iorga B, Schwanke K, Weber N, Wendland M, Greten S, Piep B, Dos Remedios CG, Martin U, Zweigerdt R, Kraft T, Brenner B. Differences in Contractile Function of Myofibrils within Human Embryonic Stem Cell-Derived Cardiomyocytes vs. Adult Ventricular Myofibrils Are Related to Distinct Sarcomeric Protein Isoforms. Front Physiol. 2018 Jan 19;8:1111. doi: 10.3389/fphys.2017.01111. PMID: 29403388; PMCID: PMC5780405.
- Weber N, Schwanke K, Greten S, Wendland M, Iorga B, Fischer M, Geers-Knörr C, Hegermann J, Wrede C, Fiedler J, Kempf H, Franke A, Piep B, Pfanne A, Thum T, Martin U, Brenner B, Zweigerdt R, Kraft T. Stiff matrix induces switch to pure β-cardiac myosin heavy chain expression in human ESC-derived cardiomyocytes. Basic Res Cardiol. 2016 Nov;111(6):68. doi: 10.1007/s00395-016-0587-9. Epub 2016 Oct 14. PMID: 27743117.
- Viereck J, Kumarswamy R, Foinquinos A, Xiao K, Avramopoulos P, Kunz M, Dittrich M, Maetzig T, Zimmer K, Remke J, Just A, Fendrich J, Scherf K, Bolesani E, Schambach A, Weidemann F, Zweigerdt R, de Windt LJ, Engelhardt S, Dandekar T, Batkai S, Thum T. Long noncoding RNA Chast promotes cardiac remodeling. Sci Transl Med. 2016 Feb 17;8(326):326ra22. doi: 10.1126/scitranslmed.aaf1475.
