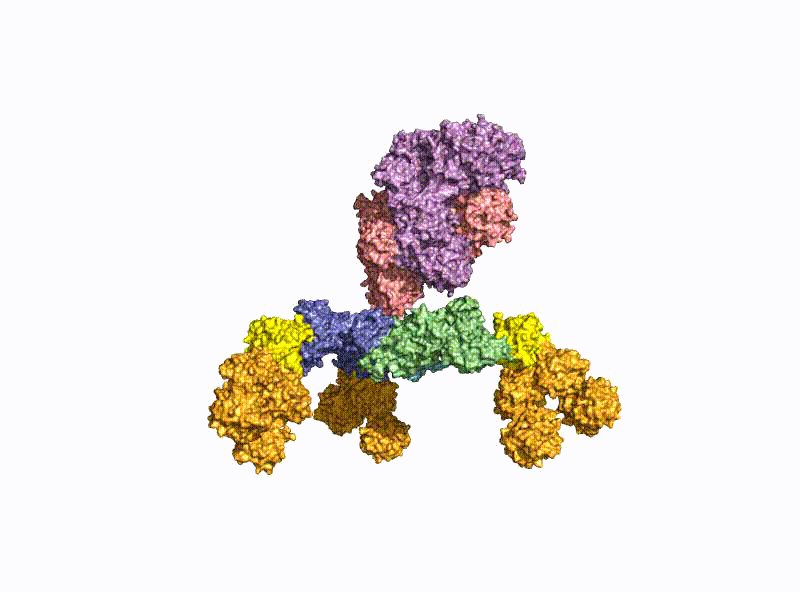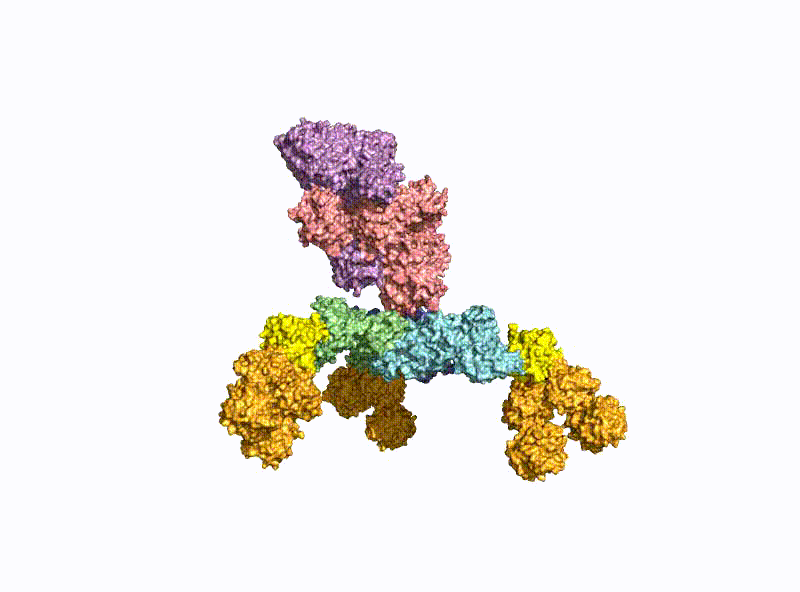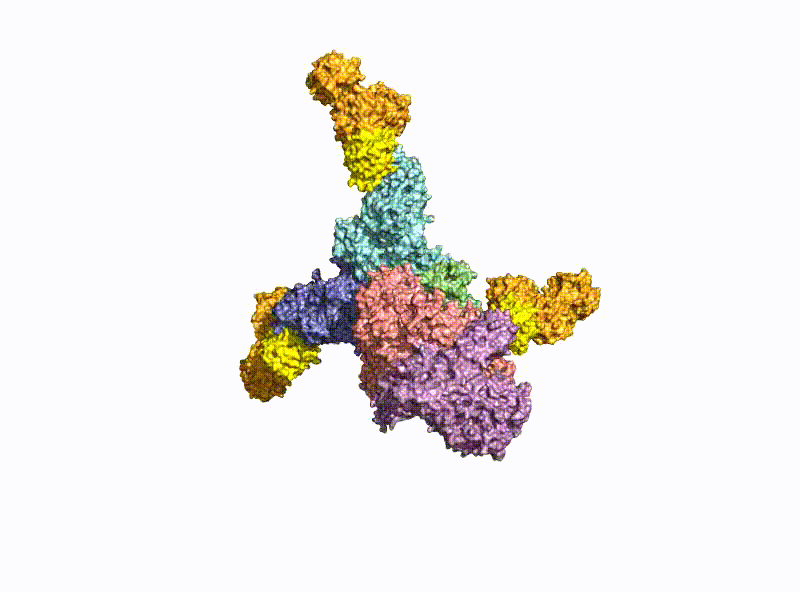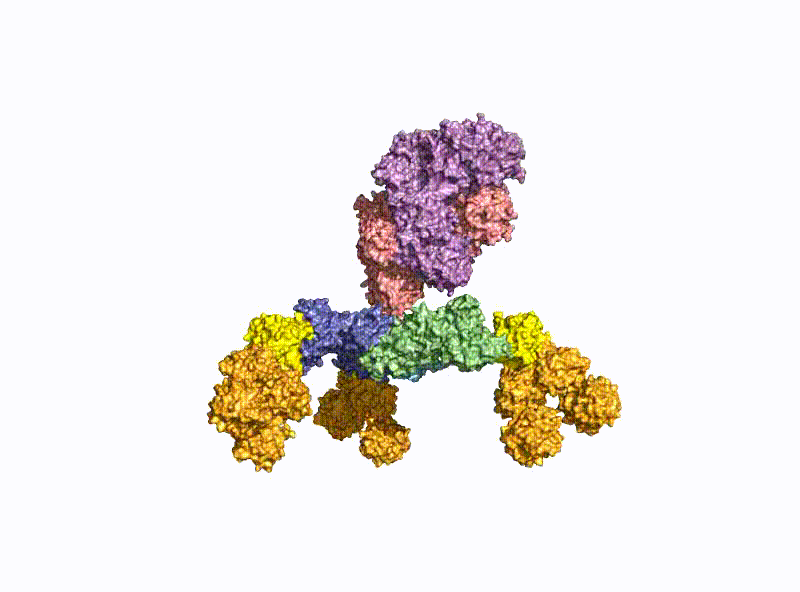Team members

from left to right
Urs von der Kammer (M.Sc.Biochemie) E-Mail
Melvin Bönninger (M.Sc.Biomedizin) E-Mail
Ariane Piwek (M.Sc.Biomedizin) E-Mail
Denise Bellmann (B.Sc.Applied Biology) E-Mail
Dr. rer. nat. Jasmin Weisemann E-Mail
Dr. rer. nat. Andreas Rummel E-Mail
No photo: Nadja Krez E-Mail
Annika Merten (M.Sc.Animal Biology and Biomedical) E-Mail
Research
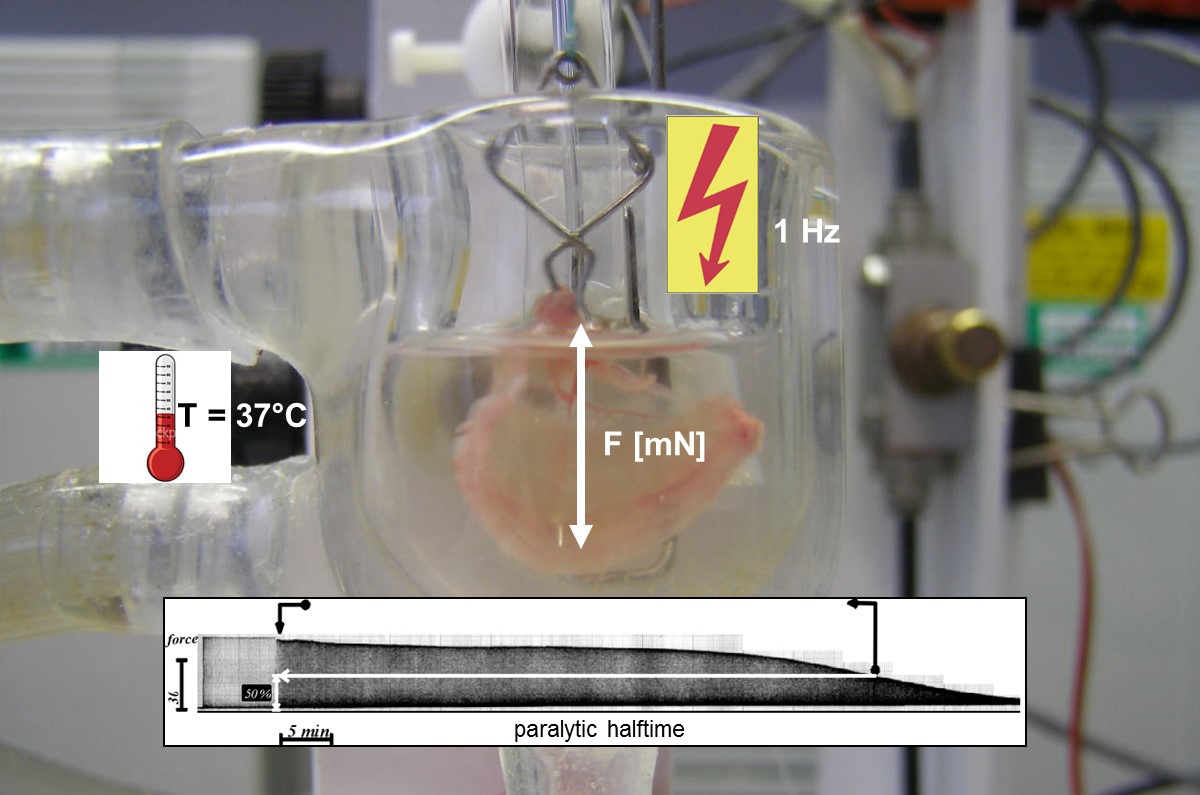
The group leader Dr. Andreas Rummel has been researching the basic mechanisms of action of clostridial neurotoxins at the Hannover Medical School (MHH) continuously since 1998 and has thus contributed to increasing our knowledge of the molecular mechanisms of action of botulinum neurotoxins (BoNTs) and their detection [1, 2]. As a result, he has currently published 50 peer-reviewed original publications and 13 review articles / book chapters, which have been cited over 3800 times (as of 2020). At the Institute for Toxicology of the MHH, he and his group have built up a high level of expertise in the field of recombinant expression of BoNTs in E. coli. Dr. Rummel was and is among other projects principal investigator (PI) in the BMBF-funded projects FuMiBoNT (Fk 031A212A; final report), SensTox (Fk 13N13794; final report) and TiViBoNT (Fk 031L0111). In these, methods for the production of bacterial protein toxins and their protein receptors were successfully researched, which can be used as reagents for detection of e.g. botulinum neurotoxins in an animal test replacement method for botulism diagnostics. In addition, the Rummel lab successfully operates an animal experiment replacement method for BoNT diagnostics, the phrenic nerve hemidiaphragm assay [3], which is also used in teaching to demonstrate the effect of licensed muscle relaxants (Fig. 1).
Botulinum Neurotoxins
The name botulinum neurotoxin (BoNT) comprises a family of more than 50 bacterial protein toxins that block the release of acetylcholine in the neuromuscular endplate and thus cause the fatal disease botulism in the event of oral intoxication. From 1897 to 1970, seven serotypes BoNT/A-G were identified which differ in the amino acid sequence from 37.2% to 69.6% [4]. The name botulinum neurotoxin (BoNT) comprises a family of more than 50 bacterial protein toxins that block the release of acetylcholine in the neuromuscular endplate and thus cause the fatal disease botulism in the event of oral intoxication. From 1897 to 1970, seven serotypes BoNT/A-G were identified which differ in the amino acid sequence from 37.2% to 69.6% [5, 6]. In October 2013, a new BoNT serotype HA, which is produced by a bivalent C. botulinum strain isolated from the stool of a patient with infant botulism, was identified [7, 8, 9]. In the meantime, further toxin genes coding for new serotypes, provisionally called BoNT/X, eBoNT/J aka BoNT/En, have been identified and characterized in part molecularly and structurally using non-toxic fragments [10, 11, 12, 13, 14].
Recently, PMP1, a BoNT-like neurotoxin that selectively targets Anopheles mosquitoes, was identified from Paraclostridium bifermentans strains collected in endemic Anopheles areas on two continents [15].
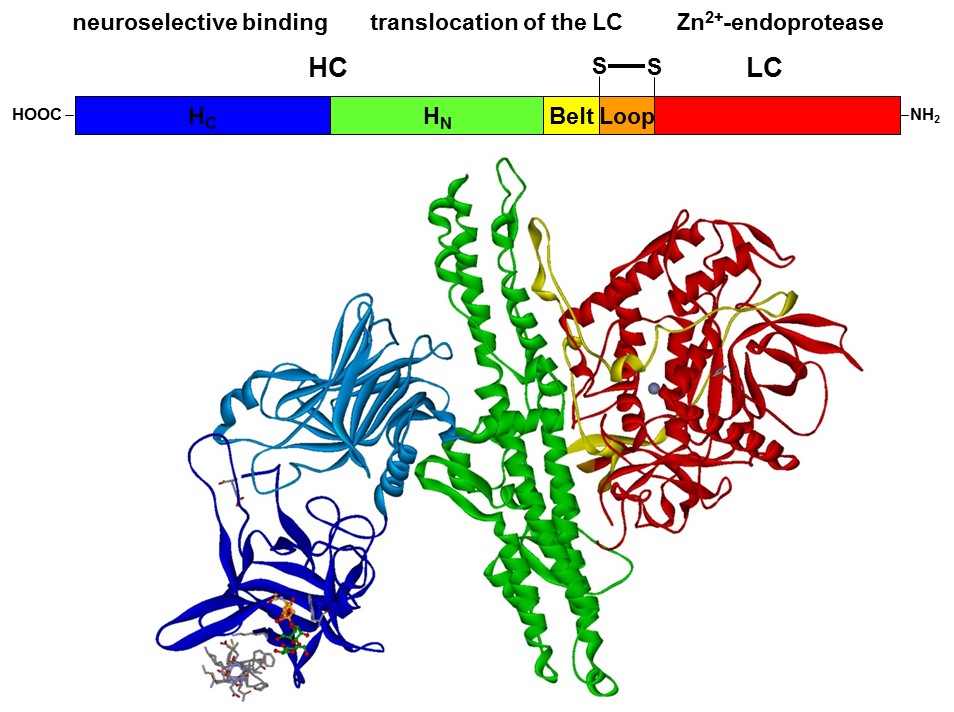
With a molecular weight of 150 kDa, the BoNTs are classic AB toxins (Fig. 2) and consist of a light chain (LC) with 50 kDa and a heavy chain (HC) with 100 kDa, which are covalently linked via a disulfide bridge. The HC is divided into a translocation domain HN and a receptor binding domain HC, each with 50 kDa.
Scientific foci
One focus of our group was and is the elucidation of the receptor entities and their interaction with binding pockets in HC. We exploit this knowledge for the development of pharmaceutically improved active ingredients, capture structures for the detection of BoNT and inhibitors for the treatment or prevention of botulism.
A second focus of our group is research into the oral uptake of BoNTs, which are some of the few proteins that survive the harsh conditions in the gastrointestinal tract unscathed. For this purpose, they form pH-dependently an acid- and protease-stable medium-sized (M-) complex with the almost equally large non-toxic non-hemagglutinin (NTNHA) (Fig. 3), whose structure was solved by our group in close collaboration with the group of R. Jin (UCI, CA) and published in Science in 2012 [18].Three other hemagglutinins (HA) of different sizes form the so-called HA complex of 510 kDa, which combines with the M complex to form the 760 kDa large BoNT (L-) complex, which is a highly potent food poison. Our group researched in detail the structure of the L-complex and the interaction of the HA-complex with sugar structures in the small intestine [17] as well as its intestinal absorption mechanism by breaking cell-cell contacts [19].
Future research fields lie in the area of the molecular characterization of new members of the BoNT family with regard to oral intake and intestinal absorption, neuronal receptor interaction and enzymatic effects in the nerve cell.
We are always looking for interested Bachelor and Master students from natural science courses as well as doctoral students in the StrucMed program.
Contact:
Dr. Andreas Rummel
E-Mail