Project presentation 2023
Single-cell transcriptomics reveals common epithelial response patterns in human acute kidney injury
Christian Hinze, Christine Kocks, Janna Leiz, Nikos Karaiskos, Anastasiya Boltengagen, Shuang Cao, Christopher Mark Skopnik, Jan Klocke, Jan-Hendrik Hardenberg, Helena Stockmann, Inka Gotthardt, Benedikt Obermayer, Laleh Haghverdi, Emanuel Wyler, Markus Landthaler, Sebastian Bachmann, Andreas C Hocke, Victor Corman, Jonas Busch, Wolfgang Schneider, Nina Himmerkus, Markus Bleich, Kai-Uwe Eckardt, Philipp Enghard, Nikolaus Rajewsky, Kai M Schmidt-Ott
Acute kidney injury is a serious complication and affects approximately 20% of hospitalized patients. In intensive care patients, up to 50% of those treated suffer acute kidney injury. The causes for this are very heterogeneous; acute kidney injury can be triggered, for example, by severe trauma and infections, complicated operations, as well as intoxication and adverse drug effects. The presence of even mild acute kidney injury is associated with increased long-term mortality. Unfortunately, there are currently no targeted kidney therapies for most forms of acute kidney injury. This is not least due to a very incomplete understanding of the exact molecular processes in the kidneys in the event of acute kidney injury.
In the 2023 Research Report, we therefore present a study by nephrologists Dr. Christian Hinze and Professor Kai Schmidt-Ott from the Clinical Department of Nephrology and Hypertension, who, in collaboration with scientists from Charité-Universitätsmedizin and the Max Delbrück Center in Berlin, have investigated the molecular processes involved in acute kidney injury. The underlying work was published in the journal Genome Medicine.
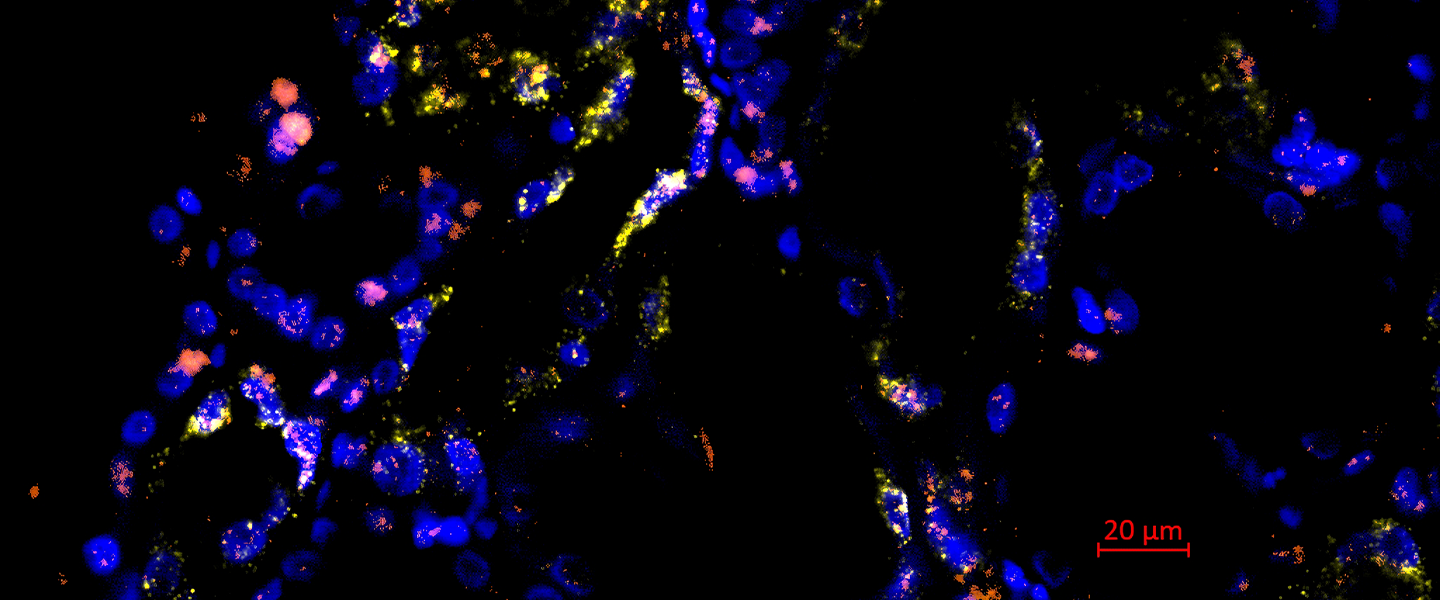
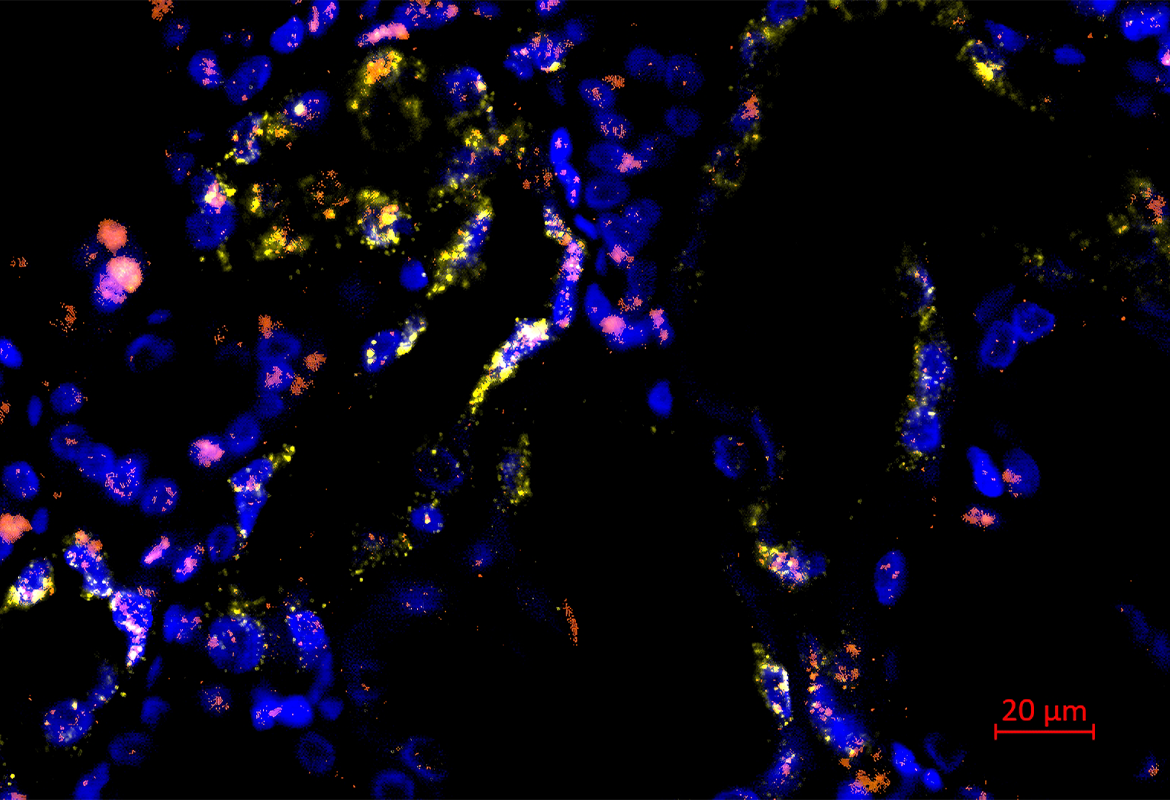
Since the kidney itself consists of a large number of different cell types, the researchers used high-resolution molecular methods of single-cell sequencing, which make it possible to examine the gene expression profile of thousands of cells individually. Kidney biopsies were obtained from patients with severe acute kidney injury who were receiving intensive care for severe respiratory infections. These were compared with healthy control kidney tissue, which was obtained from non-diseased areas of the kidneys during kidney operations, for example.
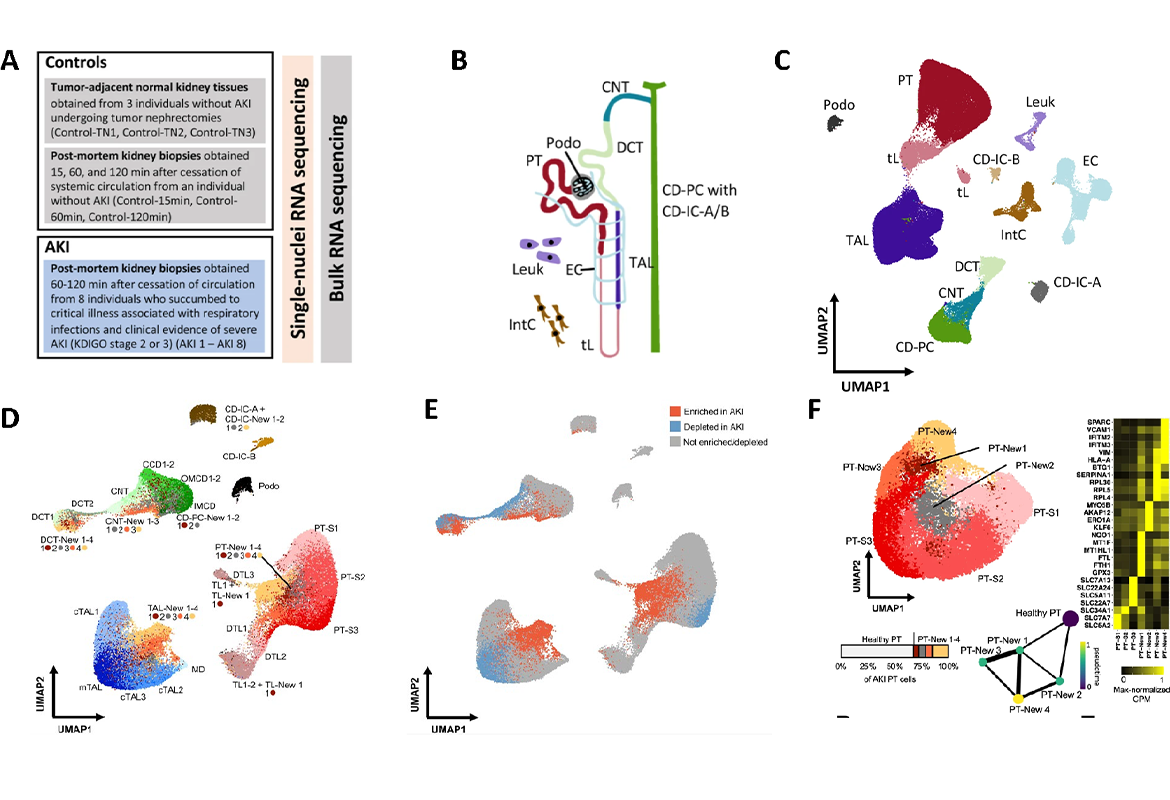
For this study, the researchers sequenced the transcriptome (i.e. the entirety of the genes transcribed in a cell) of more than 100,000 individual cells from a total of 12 patients (8 with acute kidney injury and 4 controls without acute kidney injury). Bioinformatic analyses made it possible to assign the sequenced cells to the original cell types. The analysis revealed that the gene expression of those affected by acute kidney injury differed considerably from that of healthy kidney tissue. It was seen that acute kidney injury led to the formation of new pathological cell states in cells of the renal tubule system. Interestingly, these changes appeared to occur in a similar way in different cell types of the renal tubules. The researchers were able to identify four different damage-associated cell states, each of which had defined gene expression profiles and could be validated using different methods. In addition to differences in the gene expression profile, the cell states associated with acute kidney injury apparently also differed in their potential to regenerate back into healthy kidney cells, as shown by a bioinformatic comparison with time-resolved single-cell sequencing data from a mouse model of kidney injury. Some of the damage-associated cells appear to have the potential to regenerate into healthy kidney tissue, while other cells showed an irreversible pattern of damage (see figure).
The work is one of the first studies to use single cell sequencing to investigate the diversity of cell type-specific gene expression responses to acute kidney injury in humans. The signaling pathways that are activated in the damage-associated cells provide new clues for possible therapeutic interventions. For example, new therapies could aim to inhibit the signaling pathways typical of irreversibly damaged cells.
Publication
Single-cell transcriptomics reveals common epithelial response patterns in human acute kidney injury
Hinze C, Kocks C, Leiz J, Karaiskos N, Boltengagen A, Cao S, Skopnik CM, Klocke J, Hardenberg JH, Stockmann H, Gotthardt I, Obermayer B, Haghverdi L, Wyler E, Landthaler M, Bachmann S, Hocke AC, Corman V, Busch J, Schneider W, Himmerkus N, Bleich M, Eckardt KU, Enghard P, Rajewsky N, Schmidt-Ott KM
Further information
Publisher:
President of Hannover Medical School
Prof. Dr. med. Michael P. Manns
Dean of Research at Hannover Medical School
Prof. Dr. med. Frank M. Bengel
Editing and contact person:
Reporting Department of Hannover Medical School
Alica Wollmann
Phone: 0511/ 532- 5578
E-mail: wollmann.alica@mh-hannover.de
Online implementation:
Office of the Dean of Research at Hannover Medical School
Jan Tauwaldt
and
Reporting Department of Hannover Medical School
Alica Wollmann
Phone: 0511/ 532- 5578
Research report 2023
Here you can find the research report created with the help of the Research Information System (FIS). As in previous years, we would like to take the opportunity here to explicitly present one project as a representative.
Research Information System (FIS)Here you can find further information on the Research Information System (FIS).
University bibliography- Project presentation 2022
- Project presentation 2021
- Project presentation 2020
- Project presentation 2019
- Project presentation 2018
- Project presentation 2017
- Research report 2016
- Research report 2015
- Research report 2014
- Research report 2013
- Research report 2012
- Research report 2011
- Research Report 2010
- Research Report 2009
- Research Report 2008
- Research Report 2007